In this article, I want to explore in the matter of predicting the heart disease using R Language. I want to know if there are variables that are influencing heart disease conditions. Therefore, I created a project in regards to predicting the heart disease using R Language.
In this project, the first thing to do is prepare the library that are facilitating me to do the analysis.
Import the packages
require(C50)
require(caret)
require(e1071)
require(caTools)
After importing the packages, we need to import the data that will be used for analysis. We uses heart_dataset.csv which are contains many variables related to heart disease.
Import the data
heart <- read.csv(file.choose(),sep=',')
set.seed(1234)
heart$target <- as.factor(heart$target)
vars <- c("trestbps", "chol","thalach")
str(heart[, c(vars, "target")])
Once you have imporrted the data, split the data into train and test data.
Split the data
coba <- sample(1:nrow(heart), size = 250 ,replace=TRUE)
train_data <- heart[ coba,]
test_data <- heart[-coba,]
Then, create the model using C5.0 Algorithm.
Create the model
mod <- C5.0(x = train_data[,vars],y = train_data$target)
mod
Here is the model which has been created.
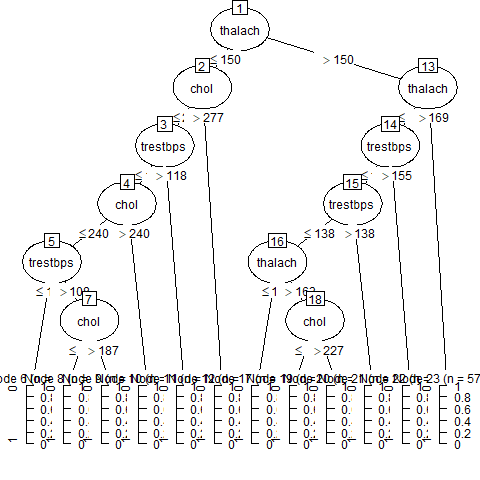
After that, we need to validate the model to know to find out whether the model made is a good model or not.
Validate the model
hasil<- predict(object=mod, newdata=test_data, type="class")
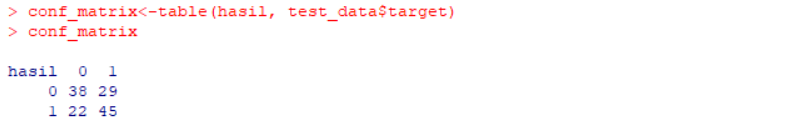
table(hasil, test_data$target)
conf_matrix<-table(hasil, test_data$target)
conf_matrix
akurasi<-round((conf_matrix[1,1]+conf_matrix[2,2])/nrow(test_data),2)*100
akurasi
error_rate<-100-akurasi
error_rate
Here are the confusion matrix and the accuracy from the model that has been created.
Confusion Matrix
Accuracy
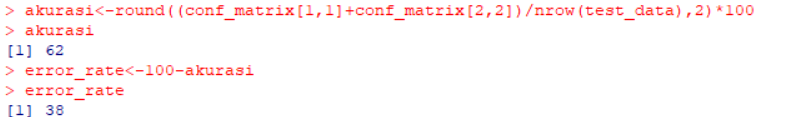
As you can see, This project resulting in an accuracy of 62% which is a fairly low level of accuracy. The accuracy is so small because it uses a categorical variables with different levels, which tends to decrease the accuracy.